Predicting genetic diversity trajectories in the Anthropocene
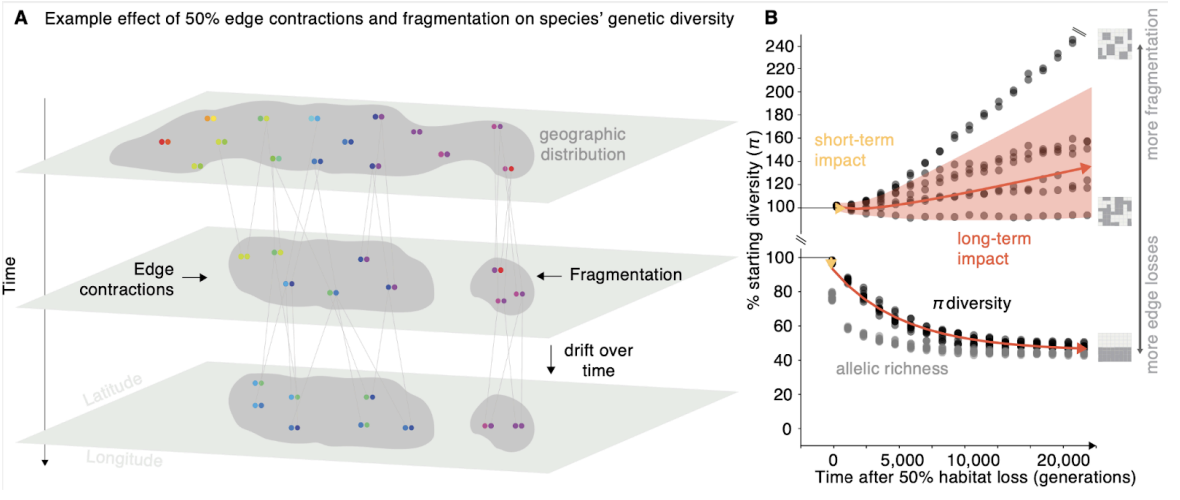
Genetic diversity is crucial for species' ability to adapt and has been targeted for protection in the UN's Global Biodiversity Framework. However, predicting genetic diversity loss is challenging. Using genetic data from 18 species, we found that genetic diversity declines slowly after habitat loss but continues for decades even if habitats are protected. Our model, combined with habitat data and conservation indicators, projects significant genetic diversity losses in 13,808 species, with short-term losses of 13-22% and long-term losses of 42-48%. These findings suggest that protecting current habitats alone is not enough to maintain genetic health, emphasizing the need for ongoing genetic monitoring and a new predictive framework for global genetic biodiversity