Phylo - a multiple sequence alignment problem
Published:
What is MSA (Multiple Sequence Alignment?)
One of the most fundamental and powerful techniques in molecular biology is our ability to compare genomes across species, in an attempt to decipher and understand our complex genome. The act of aligning sequences allows us to identify regions of similarity across sequences, which could be brought about by functional, structural or evolutionary relationships betweeen the sequences.
The Problem
The problem stems from many multiple sequence alignment algorithms being computationally complex heuristics. However, the use of current multiple sequence alignment heuristics do not guarantee a global optimized solution and becomes increasingly computationally expensive in proportion to genome size.
The Solution
Utilizing the computational ability of the human mind, tuned for instinctive pattern recognition, we extended the multiple sequence alignment problem to people in the form of an interactive platform. The data extracted from this platform can then be further utilized to optimize for global alignments such as feature selection w.r.t the original puzzles.
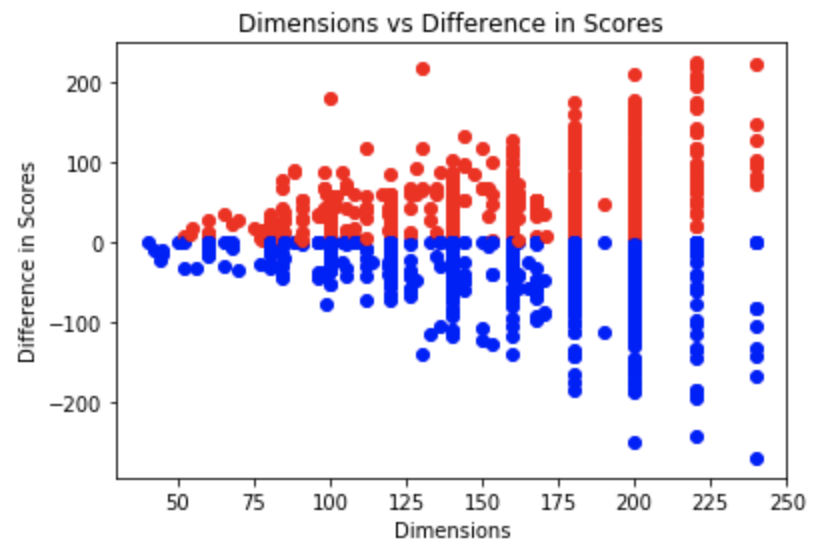
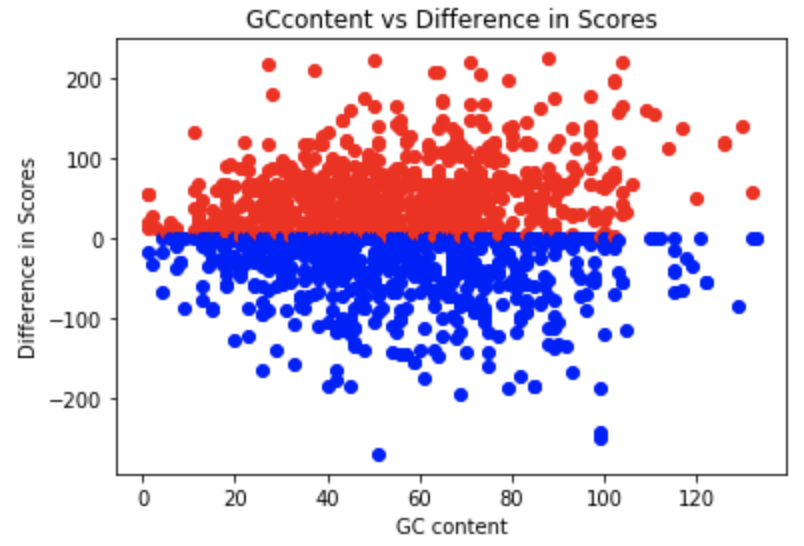
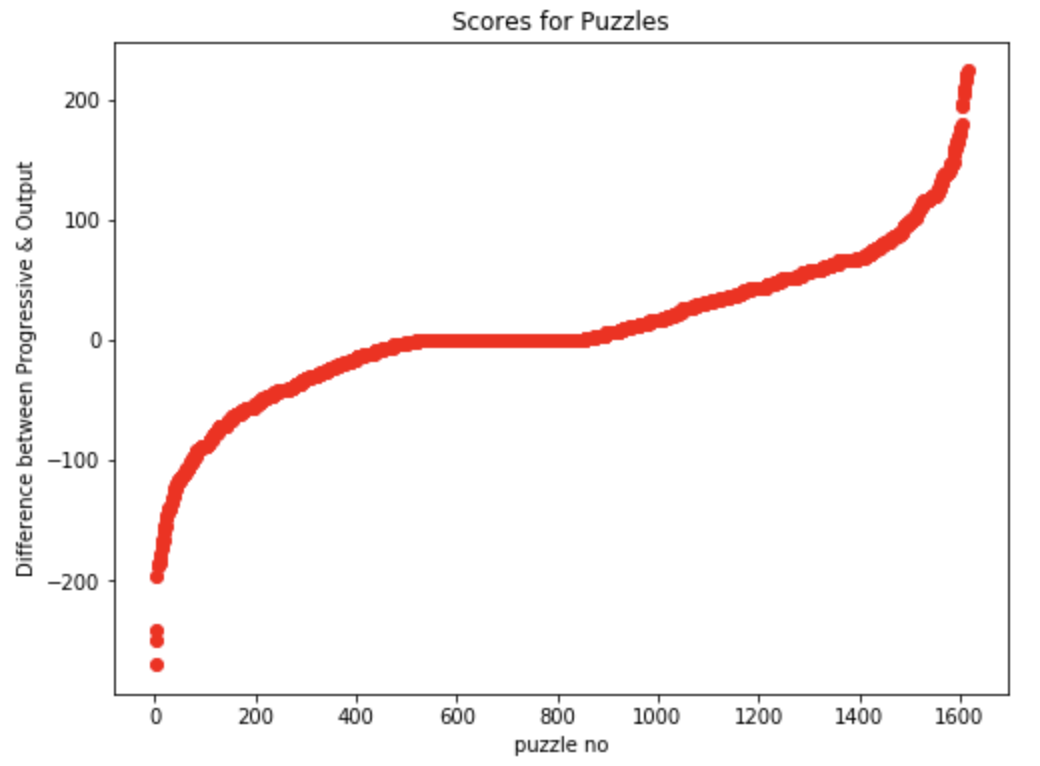
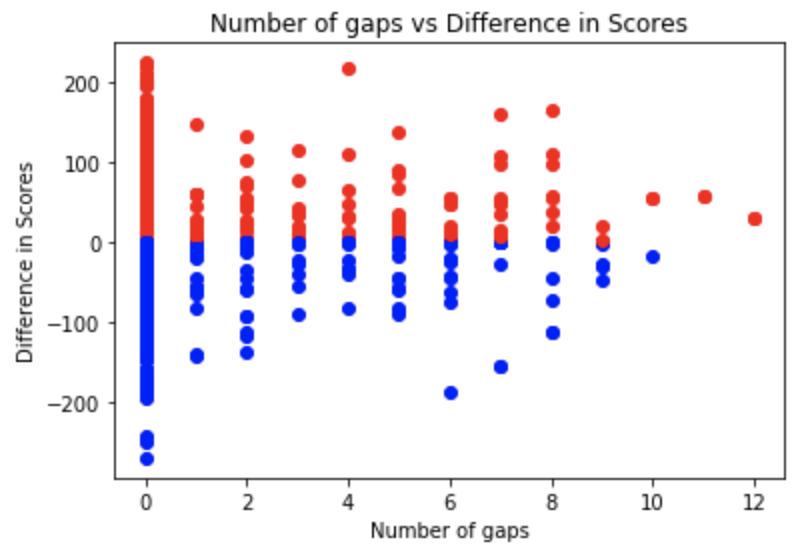
Fig 1-4. Data represention of feature selection for Phylo Puzzles
